Two publications from Dr. Pratik Shah's lab at IEEE ICMLA
December 18, 2018 - December 19, 2018
8:00am - 7:20pm ET
Orlando, FL
17th IEEE International Conference on Machine Learning and Applications (ICMLA 2018) aims to bring together researchers and practitioners to present their latest achievements and innovations in the area of machine learning. Contributions describing machine learning techniques applied to real-world problems and interdisciplinary research involving machine learning, in fields like medicine, biology, industry, manufacturing, security, education, virtual environments, games, will be presented as oral presentations (full papers) and posters (short papers). Conference content will be submitted for inclusion into IEEE Xplore as well as other Abstracting and Indexing (A&I) databases.
2018 Conference paper acceptance rate: 14%
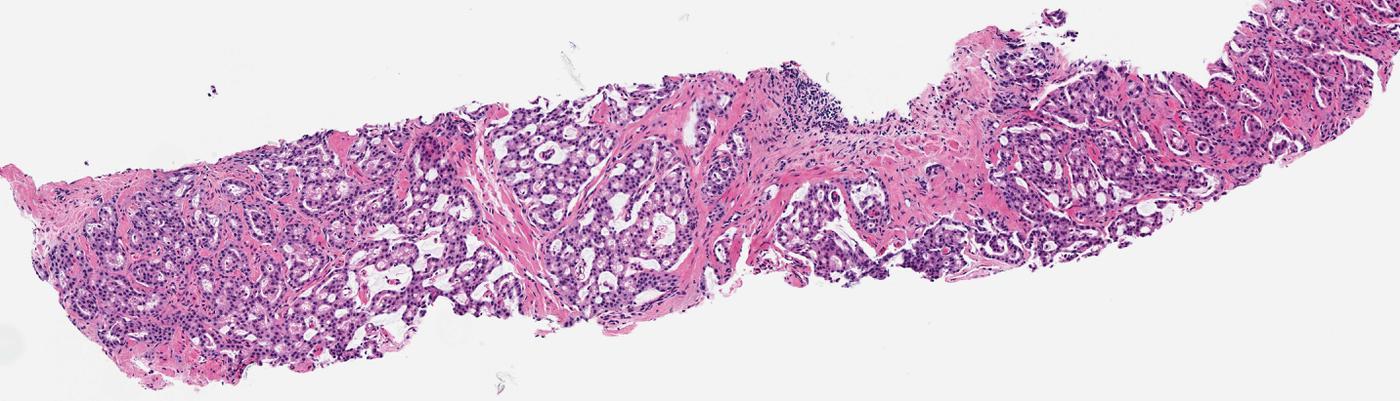
Paper 1: Computational Histological Staining and Destaining of Prostate Core Biopsy RGB Images with Generative Adversarial Neural Networks
Aman Rana, Gregory Yauney, Alarice Lowe, Pratik Shah
12/18/18, 5:20-7:20pm, short paper and poster
Abstract: Histopathology tissue samples are widely available in two states: paraffin-embedded unstained and non-paraffin-embedded stained whole slide RGB images (WSRI). Hematoxylin and eosin stain (H&E) is one of the principal stains in histology but suffers from several shortcomings related to tissue preparation, staining protocols, slowness and human error. We report two novel approaches for training machine learning models for the computational H&E staining and destaining of prostate core biopsy RGB images. The staining model uses a conditional generative adversarial network that learns hierarchical non-linear mappings between whole slide RGB image (WSRI) pairs of prostate core biopsy before and after H&E staining. The trained staining model can then generate computationally H&E-stained prostate core WSRIs using previously unseen non-stained biopsy images as input. The destaining model, by learning mappings between an H&E stained WSRI and a non-stained WSRI of the same biopsy, can computationally destain previously unseen H&E-stained images. Structural and anatomical details of prostate tissue and colors, shapes, geometries, locations of nuclei, stroma, vessels, glands and other cellular components were generated by both models with structural similarity indices of 0.68 (staining) and 0.84 (destaining). The proposed staining and destaining models can engender computational H&E staining and destaining of WSRI biopsies without additional equipment and devices.
*Paper 2: Machine Learning Algorithms for Classification of Microcirculation Images from Septic and Non-Septic Patients
*Perikumar Javia, Aman Rana, Nathan Shapiro, Pratik Shah
12/19/18, 2:00-3:20pm, full paper and invited talk
Abstract: Sepsis is a life-threatening disease and one of the major causes of death in hospitals. Imaging of microcirculatory dysfunction is a promising approach for automated diagnosis of sepsis. We report a machine learning classifier capable of distinguishing non-septic and septic images from dark field microcirculation videos of patients. The classifier achieves an accuracy of 89.45%. The area under the receiver operating characteristics of the classifier was 0.92, the precision was 0.92 and the recall was 0.84. Codes representing the learned feature space of trained classifier were visualized using t-SNE embedding and were separable and distinguished between images from critically ill and non-septic patients. Using an unsupervised convolutional autoencoder, independent of the clinical diagnosis, we also report clustering of learned features from a compressed representation associated with healthy images and those with microcirculatory dysfunction. The feature space used by our trained classifier to distinguish between images from septic and non-septic patients has potential diagnostic application.