Uncertainty-Quantified Deep Learning for Image Segmentation of Prostate and Skin Cancer
Motivation
Clinical deployment of deep learning models for medical image segmentation is limited by the absence of uncertainty quantification and performance feedback. Dr. Pratik Shah’s lab developed a workflow that estimates Dice coefficients and prediction uncertainty in individual regions of medical images to support review and interpretation. Two projects—focused on prostate biopsy slides and dermoscopy images—report statistical methods and visual tools that use region-level uncertainty to evaluate and predict deep learning model segmentation performance.
Technical Summary
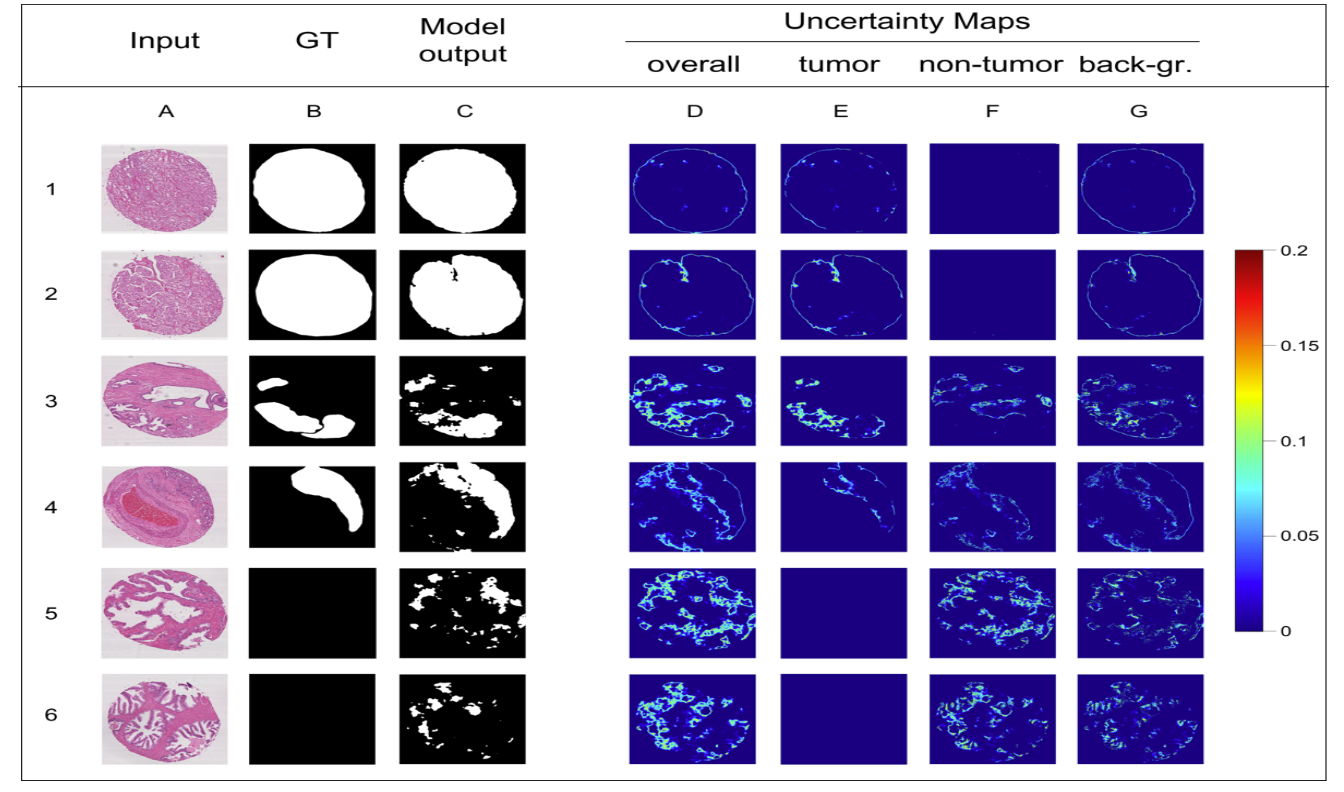
This research project introduces regression-based models to estimate the Dice performance of deep learning segmentation tasks using Monte Carlo Dropout (MCD) and Bayes-by-Backprop (BBP). Two categories of images—prostate cancer biopsies and dermoscopic images of skin lesions—were used to develop deep learning models trained from scratch or with transfer learning. Uncertainty maps were computed at pixel level and aggregated across clinical regions of interest (ROIs). Region-based uncertainty measures were correlated with Dice scores, and linear models were developed to predict segmentation quality. These methods allow automatic estimation of segmentation reliability from a single image and facilitate identification of predictions for review.
FAQ
What is the value proposition of this work?
This work introduces statistical methods to estimate segmentation performance of deep learning models using uncertainty measurements. The approach uses multiple clinical regions of interest within a medical image to produce uncertainty maps and applies linear regression to predict Dice scores for individual images. The process helps assess prediction quality from previously unseen inputs without requiring ground truth, making it suitable for integration into clinical workflows.
What were the key findings from this work?
- Uncertainty estimates are inversely correlated with Dice performance across both prostate and skin datasets.
- Algorithms that enables unsupervised Dice prediction using image-level uncertainty without requiring ground-truth labels.
- Dice scores can be predicted using statistical models trained on region-based uncertainty derived from MCD or BBP methods.
- Segmentation uncertainty can be localized within clinically relevant regions such as tumor or lesion areas.
What are the main outcomes and the meaning of this work for deep learning research and clinical applications?
This study provides methods for evaluating segmentation performance without labeled data, enabling integration of deep learning into real-world settings where validation labels are not available. The work supports deployment of segmentation models in histopathology and dermatology by identifying uncertain predictions that require clinician review. The proposed uncertainty framework also supports selection of reliable images for training and performance tuning.
What are the next steps?
Ongoing work includes generalizing the uncertainty framework to other imaging modalities and clinical domains. Future studies will simulate noise conditions and evaluate robustness under adversarial inputs. Collaborative studies are planned with hospitals and regulatory partners to prospectively validate the model in clinical workflows and explore its utility in decision support systems.
What are the limitations and future growth areas of this research?
The models were trained and validated on selected datasets with predefined annotations, which may not generalize to other labeling schemes or imaging conditions. Annotations may contain coarse labels or background structures that affect uncertainty estimation. Future improvements will include more detailed clinical annotations and expanded datasets across organ systems.
Publications
- Prostate cancer segmentation study
Deep learning with uncertainty quantification for predicting the segmentation dice coefficient of prostate cancer biopsy images. 2024 IEEE International Conference on Machine Learning and Applications (ICMLA). [https://doi.org/10.1109/ICMLA61862.2024.00178].
2. Skin lesion segmentation study
Uncertainty quantified deep learning and regression analysis framework for image segmentation of skin cancer lesions. 2024 IEEE International Conference on Machine Learning and Applications (ICMLA). [https://doi.org/10.1109/ICMLA61862.2024.00080].
Contributors and coauthors
Sambuddha Ghosal & El Houcine El Fatimi: Postdoc associates
Audrey Xie: Undergraduate student
Pratik Shah^: Principal investigator (^Senior and corresponding author).
