Deep Learning to Develop and Analyze Computational Hematoxylin and Eosin Staining of Tissue Images for Digital Biopsies
Staining of tissues sections using chemical and biological dyes has been used for over a century for visualizing various tissue types and morphologic changes associated with cancer and other disorders for contemporary clinical diagnosis. This staining procedure often results in loss of irreplaceable tissue specimen and delays diagnoses. Other key challenges include sampling time, which can limit the amount of tissue that can be stained owing to time and cost involved, resulting in evaluation of only three 4-μm sections of tissue to represent a 1-mm diameter core. Irreversible dye staining of tissues leads to loss of precious biopsy samples that are no longer available for biomarker testing.
Dr. Shah’s lab has previously described generative computational methods that use neural networks that rapidly stain photographs of non-stained tissues, providing physicians timely information about the anatomy and structure of the tissue. The lab also reported a “computational destaining” method that can remove dyes and stains from photographs of previously stained tissues, allowing reuse of patient samples. However, studies testing operational feasibility and validation of results obtained by these generative neural network models and machine learning algorithms in controlled clinical trials or hospital studies for virtual staining of whole-slide pathology images did not exist, precluding clinical adoption and deployment of these systems.
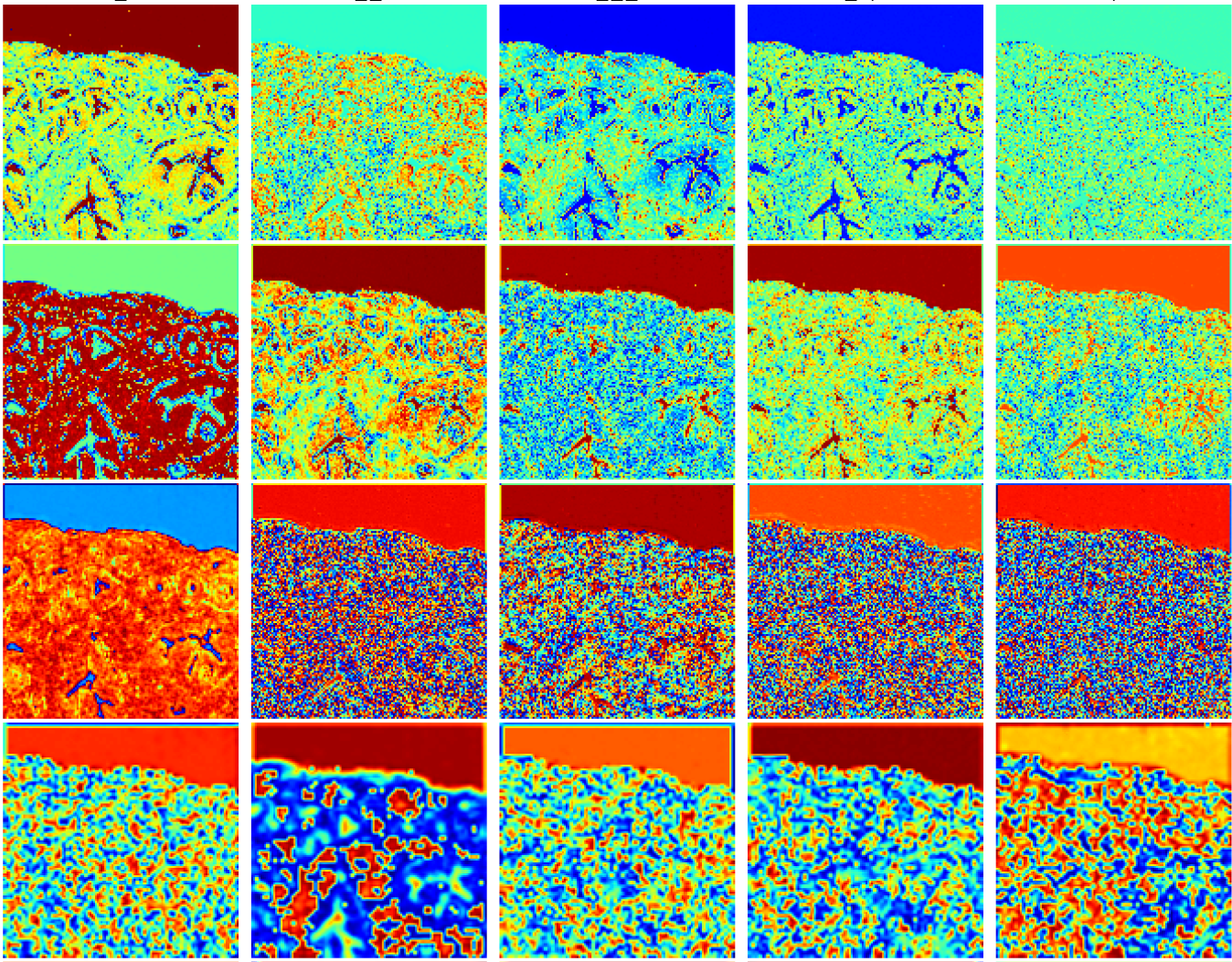
In this study led by Dr. Shah, in collaboration with Stanford University School of Medicine and Harvard Medical School, several novel mechanistic insights and methods to facilitate benchmarking and clinical and regulatory evaluations of generative neural networks and computationally H&E stained images were reported. Specifically, high fidelity, explainable, and automated computational staining and destaining algorithms to learn mappings between pixels of nonstained cellular organelles and their stained counterparts were trained. A novel and robust loss function was devised for the deep learning algorithms to preserve tissue structure. The study communicated that virtual staining neural network models were generalizable to accurately stain previously unseen images acquired from patients and tumor grades not part of training data. Neural activation maps in response to various tumors and tissue types were generated to provide the first instance of explainability and mechanisms used by deep learning models for virtual H&E staining and destaining. And image processing analytics and statistical testing were used to benchmark the quality of generated images. Finally, the computationally stained images were evaluated by multiple pathologists for prostate tumor diagnoses and clinical decision-making.
FAQ
-
What is the value proposition of this work?
Can deep learning systems perform Hematoxylin and Eosin (H&E) staining and destaining, and are the virtual core biopsy samples generated by them as valid and interpretable as their real-life unstained and H&E dye–stained counterparts. And how can clinicians, researchers and regulatory agencies understand and authenticate them before integration into healthcare settings.
-
What are key findings from this work?
In this novel cross-sectional diagnostic study, deep learning models [conditional Generative Adversarial Neural Networks (cGANS)] were trained using nonstained prostate core biopsy images to generate computationally H&E stained images. The study also communicated computational H&E destaining cGANS that could reverse the process. A single blind prospective study computed approximately 95% pixel-by-pixel overlap among prostate tumor annotations provided by 5 board certified pathologists on computationally stained images, compared with those on H&E dye–stained images. The study also reports novel visualization and explanation of neural network kernel activation maps during H&E staining and destaining by cGANs. High similarities between kernel activation maps of computationally and H&E-dye-stained images (mean-squared errors <0.0005) provide additional mathematical and mechanistic validation of the staining system.
-
What are the main outcomes and the meaning of this work for deep learning research and clinical applications?
Methods for detailed computer vision image analytics, visualization of trained cGAN model outputs, and clinical evaluation of virtually stained images were developed. The main outcome was interpretable deep learning models and computational H&E-stained images that achieved high performance in these metrics. The findings of this study indicate that whole slide nonstained microscopic images of prostate core biopsy, instead of tissue samples, can be integrated with deep learning algorithms to perform computational H&E staining and destaining for rapid and accurate tumor diagnosis by human physicians. These findings also indicate that computational H&E staining of native unlabeled RGB images of prostate core biopsy could reproduce Gleason grade tumor signatures that can be easily assessed and validated by clinicians. Methods for benchmarking, visualization, and clinical validation of deep learning models and virtually H&E-stained images communicated in this study have applications in clinical informatics and computer science research. Clinical researchers may use these systems for early indications of possible abnormalities in native nonstained tissue biopsies prior to histopathological workflows.
-
What is the significance of this work for integration of Software as a Medical Device (SaMD) into clinical practice ?
United States Food and Drug Administration is in the process of making key determinations for regulatory strategies provided by SaMD for health care decisions to treat, diagnose, drive, support or inform clinical management. By describing explainable algorithms and quantitative methods that can consistently, rapidly, and accurately perform computational staining and destaining of prostate biopsy digital slides, this study communicates a detailed method and process that may be useful to generate evidence for clinical and regulatory authentication of SaMD. Dr. Shah has ongoing collaborations with regulatory agencies, academic hospitals and foundations for prospective validation and randomized clinical trials.
-
What are the next steps?
The clinical outcomes from this study are limited to the evaluation of prostate core biopsies as a representative tissue type, but the methods and approach should generalize to other tissue biopsy evaluations. Application to other tumor types within core biopsies or to resection specimens of prostate cancer or other conditions will be evaluated in future work. Greater numbers of virtually stained H&E images sourced from larger pools of patients are needed before prospective clinical evaluation of models described in this study can begin.
Contributors and coauthors
Aman Rana*, MS: Massachusetts Institute of Technology
Alarice Lowe*, MD: Stanford University School of Medicine.
Marie Lithgow, MD: Boston University School of Medicine, VA Boston Healthcare
Katharine Horback, MD: Harvard Medical School, Brigham and Women’s Hospital
Tyler Janovitz, MD: Harvard Medical School, Brigham and Women’s Hospital
Annacarolina Da Silva, MD: Harvard Medical School, Brigham and Women’s Hospital
Harrison Tsai, MD: Harvard Medical School, Brigham and Women’s Hospital
Vignesh Shanmugam, MD: Harvard Medical School, Brigham and Women’s Hospital
Akram Bayat, PhD: Massachusetts Institute of Technology
Pratik Shah, PhD^: Massachusetts Institute of Technology
*: Contributed equally
^: Senior author supervising research.

Dr. Pratik Shah
Faculty Member
Other Contributors
Contributors and coauthors
Aman Rana*, MS: Massachusetts Institute of Technology
Alarice Lowe*, MD: Stanford University School of Medicine.
Marie Lithgow, MD: Boston University School of Medicine, VA Boston Healthcare
Katharine Horback, MD: Harvard Medical School, Brigham and Women’s Hospital
Tyler Janovitz, MD: Harvard Medical School, Brigham and Women’s Hospital
Annacarolina Da Silva, MD: Harvard Medical School, Brigham and Women’s Hospital
Harrison Tsai, MD: Harvard Medical School, Brigham and Women’s Hospital
Vignesh Shanmugam, MD: Harvard Medical School, Brigham and Women’s Hospital
Akram Bayat, PhD: Massachusetts Institute of Technology
Pratik Shah, PhD^: Massachusetts Institute of Technology
*: Contributed equally
^: Senior author